Forensic genetics is the use of genetic tools and scientific methods to solve legal cases, both criminal and civil. The principle of Locard’s Exchange suggests that every contact leaves a trace, making any evidence crucial in forensic analysis. Biological evidence found at crime scenes can include cellular material or cell-free DNA, and as genetic technologies have advanced, these methods have been applied to both human and non-human genetic analyses. While these techniques can be used for any genome, the existence of databases and standard guidelines has made human DNA typing the preferred method.
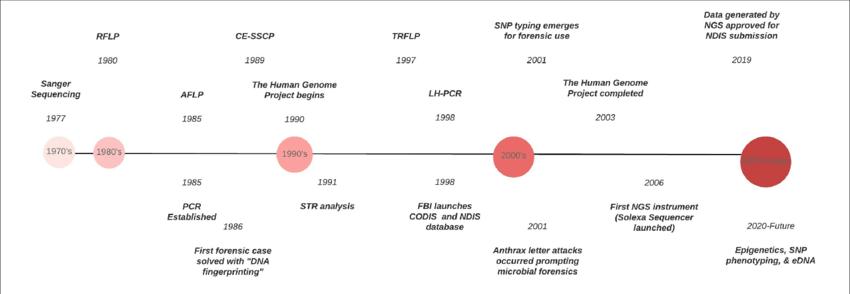
The rapid progress of forensic DNA typing can be attributed to the numerous advancements in molecular biology technologies that have occurred within a relatively short timeframe.
Overview of Forensic DNA Analysis
The techniques used for DNA fingerprinting include Restriction Fragment Length Polymorphism (RFLP) and PCR-based Variable Number Tandem Repeat (VNTR) or Short Tandem Repeat (STR) determination. While RFLP was the first approach used, it is not commonly used due to its time-consuming nature and requirement for high-quality DNA.
VNTR and STR have since been developed and are more commonly used due to their sensitivity and less time-consuming procedures. Both methods use PCR to amplify a small amount of DNA. VNTR is composed of 17-19bp repeat sequences while STR consists of 2-4bp repeat sequences.
Several different STRs are amplified by PCR and analyzed by DNA sequencing, which is currently the most commonly used method in forensic science. However, it requires expensive facilities and precision techniques.
Organization of DNA into Chromosomes
In every human nucleated cell, there are two complete copies of the genome. The human genome comprises around 3.2 billion base pairs (BPs) of information, which are arranged into 23 pairs of chromosomes. Each individual inherits one set of chromosomes from each parent, resulting in a total of 46 chromosomes.
The human genome can be classified into different types based on its structure and function.
- Firstly, there are regions of DNA that encode and regulate protein synthesis, which are called genes. The human genome is estimated to contain between 20,000 to 25,000 genes, which make up only 1.5% of the genome.
- Secondly, there are noncoding regions of genetic sequence that make up 23.5% of the genome. These regions do not encode proteins but instead play a role in regulating gene expression through enhancers, promoters, repressors, and polyadenylation signals.
- Finally, approximately 75% of the genome is extragenic DNA, which is composed of repetitive DNA. This includes 50% interspersed repeats, which consist of short interspersed elements, long interspersed elements, long terminal repeats, and DNA transposons, and 45% tandem repeats, which consist of satellite DNA, minisatellite DNA, and microsatellite DNA.
Basic Steps in Characterization of DNA Profiling and Analysis
The four basic steps involved in DNA Profiling include,
- DNA extraction
- DNA quantification
- DNA amplification
- Detection of the DNA-amplified products
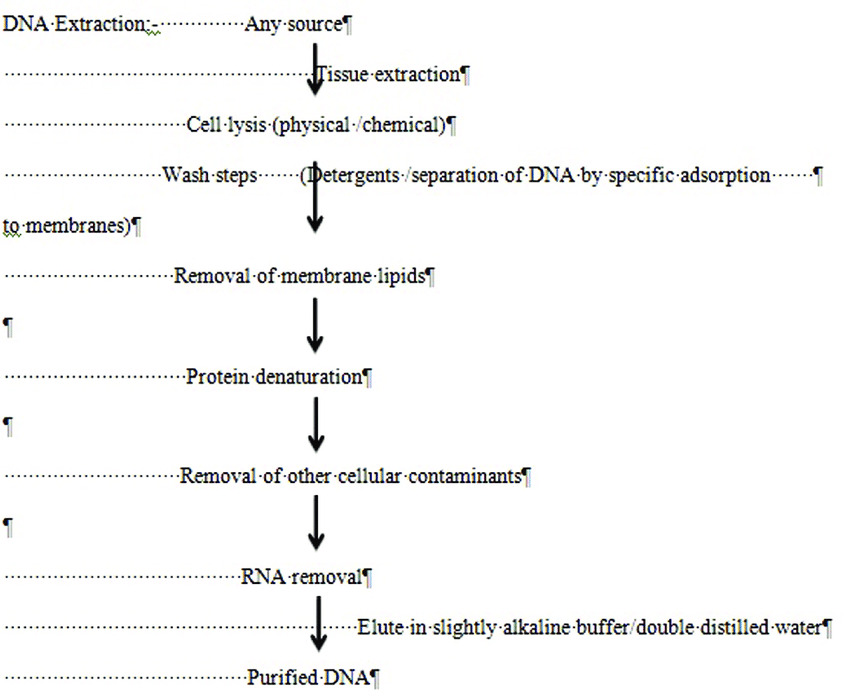
There are various methods of extraction as mentioned below, though commonly used are the Chelex-100 method, silica-based DNA extraction, and phenol–chloroform method.
- Chromatography-based DNA extraction method.
- Ethidium bromide–cesium chloride (EtBr-CsCl) gradient centrifugation method.
- Alkaline extraction method.
- Silica matrices method.
- Salting-out method.
- Cetyltrimethylammonium bromide (CTAB) extraction method.
- Sodium dodecyl sulfate (SDS)-proteinase K method.
- Silica column-based DNA extraction method.
- Cellulose-based paper method.
- Chelex-100 extraction method.
DNA Quantification
It is important to measure the amount and quality of DNA extract after DNA extraction. This is because the correct amount of DNA is crucial for obtaining the best quality results in PCR within a short time frame. If too little or too much DNA is added, it can lead to a profile that is difficult or impossible to interpret. Therefore, DNA quantification is essential to ensure optimal PCR results.
DNA quantification can be classified as follows:
- Nonnucleic acid-based quantification methods.
- Microscopic and macroscopic examination.
- Chemical and immunological methods.
- DNA-based total genomic methods.
- Intact and degraded DNA–UV spectrometry.
- PicoGreen homogenous microtiter plate assays.
- Intact vs degraded DNA–agarose gel electrophoresis.
- Real-time PCR, DNA-based target-specific methods.
- Human total autosomal DNA.
- Y chromosome DNA, mitochondrial DNA (mt-DNA), Alu repeat real-time PCR.
- Multiplex real-time PCR.
- End-point PCR DNA quantification and alternative DNA detection methods.
- RNA-based quantification.
DNA Amplification
There are different techniques for amplifying DNA and RNA, but PCR and reverse transcription-PCR is the most widely used methods.
- PCR allows the amplification of specific regions of DNA using cycling stages of denaturation, annealing, and extension.
- It can amplify a single DNA molecule to billions of copies within 30 cycles.
- The normal range of the PCR cycle is between 28 and 32, but it can be increased to 34 cycles for low DNA concentrations.
- Other amplification methods include nucleic acid sequence-based amplification, strand displacement amplification, recombinase polymerase amplification, strand invasion-based amplification, multiple displacement amplification, and hybridization chain reaction. After DNA amplification, the final step is to detect the amplified products.
Detection of the DNA-Amplified Products
The following methods are used in forensic human identification:
- Autosomal short-tandem repeat (STR) profiling
- Analysis of the Y chromosome
- Analysis of mt-DNA.
- Autosomal single-nucleotide polymorphism (SNP) typing.
Autosomal STR Profiling
The process of STR profiling involves amplifying specific regions of DNA containing STRs through polymerase chain reaction (PCR) and then analyzing the resulting DNA fragments using capillary electrophoresis.
By comparing the number and size of the amplified STR fragments between individuals, forensic scientists can differentiate between individuals with high accuracy.
STR profiling has a wide range of applications in forensic investigations, including paternity/maternity testing, rape perpetrators’ identification, kinship testing, and disaster victim identification in paternity/ maternity testing.
For example, STR profiling can determine the biological relationship between individuals by comparing their respective STR profiles. Similarly, in criminal investigations, forensic scientists can use STR profiling to match DNA evidence collected from crime scenes to suspects or their relatives.
Due to the high degree of accuracy and reproducibility of STR profiling widely accepted by both professionals and the general public as an important tool in criminal justice and human identification.
Its application has led to the resolution of many criminal cases and helped to identify victims of disasters, such as natural disasters or mass casualties resulting from terrorist attacks.
Analysis of the Y Chromosome
Biologically a male individual has 1 Y chromosome and contains 55 genes. Due to this unique feature, analysis of the Y chromosome is done in crime cases.
Application of Y chromosome in forensic medicine:
- The Y chromosome is found in males, so investigators look for it at crime scenes to help identify male suspects.
- When investigating cases involving mixtures of male and female body fluids, such as sexual assault or rape, analyzing the Y-STR component can provide more information about the male component.
- Some rapists who have undergone vasectomies or are azoospermic may not leave traces of semen, making it difficult to find spermatozoa under a microscope. In such cases, Y-STR profiling can be used in identifying the accused person.
Analysis of Mitochondrial DNA (mt-DNA)
Mitochondrial DNA (mt-DNA) is inherited exclusively from the mother, so all individuals in a matrilineal family have the same haplotype. The abundance of mt-DNA, with 200 to 1,700 copies per cell, makes it more likely to survive compared to nuclear DNA. The advantages of mt-DNA analysis include its ability to be used on severely degraded or old biological samples, as well as those with low amounts of DNA such as hair shafts.
Autosomal Single-Nucleotide Polymorphism (SNP) Typing
It has a lower heterozygosity than Short Tandem Repeat (STR) analysis. However, SNP typing has an advantage over STR analysis because the DNA template size can be as small as 50 base pairs (BPs), while STR analysis requires a template size of 300 BPs to obtain good profiling. This makes SNP typing a valuable tool for analyzing degraded samples, such as those found in the aftermath of the 2001 World Trade Center disaster, where SNP typing was used to identify victims.
Summary of the Importance of PCR in Forensic DNA Analysis
Polymerase Chain Reaction (PCR) is a critical technique in forensic DNA analysis. PCR allows the amplification of small amounts of DNA to create enough material for analysis. PCR is used in a variety of applications, including Short Tandem Repeat (STR) analysis, Mitochondrial DNA (mt-DNA) analysis, and Single-Nucleotide Polymorphism (SNP) typing. PCR is essential in forensic DNA analysis, as it enables the analysis of minute or degraded samples, such as those found at crime scenes. This technology has revolutionized forensic DNA analysis and has become an indispensable tool in forensic investigations.
References
- Past, Present, and Future of DNA Typing for Analyzing Human and Non-Human Forensic Samples Article in Frontiers in Ecology and Evolution • March 2021 DOI: 10.3389/fevo.2021.646130
- Wyman A R, White R (1980), “Highly polymorphic locus in human DNA. Proceedings of the National Academy of Sciences of the United States of America, 77(11), 6754-6758.
- Jeffreys A J, Wilson V, Thein S L (1985) Individual-specific ‘fingerprints’ of human DNA. Nature, 316(6023), 76-79.,
- Edwards A, Civitello A, Hammond H A, Caskey C T (1991) DNA typing and genetic mapping with trimeric and tetrameric tandem repeats. American Journal of Human Genetics, 49(4), 746- 756.
- International Human Genome Sequencing Consortium, “Finishing the euchromatic sequence of the human genome”, Nature 2004;431(7011):931–945.
- Goodwin W, Linacre A, Hadi serine,” An Introduction to Forensic Genetics. Chichester”, John Wiley & Sons; 2011
- Hui X, Liqun X, Jiayi cysteine, “Method for rapidly extracting nucleic acid from the biological sample”, 2014. China.
- Lee SB, McCord B, Buel glutamic acid, “Advances in forensic DNA quantification: a review. Electrophoresis”, 2014;35(21-22):3044–3052.
- Saiki RK, Gelfand DH, Stoffel S, et al. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 1988; 239 (4839):487–491.
- Gill P. Application of low copy number DNA profiling. Croat Med J 2001;42(03):229–232
- Walker FM, Hsieh K. Advances in directly amplifying nucleic acids from complex samples. Biosensors (Basel) 2019;9(04):117
- Yoshida K, Yayama K, Hatanaka A, Tamaki K. Efficacy of extended kinship analyses utilizing commercial STR kit establishing personal identification. Leg Med (Tokyo) 2011; 13(01):12–15
- Brenner CH, Weir BS. Issues and strategies in the DNA identification of World Trade Center victims. Theor Popul Biol 2003;63(03):173–178